Advancing AAV analytics: filling a critical gap in full-length transgene detection
Recombinant adeno-associated viruses (rAAVs) are fundamental to gene therapy, but a persistent challenge limits their full potential: most viral particles lack the full, intact therapeutic payload. Truncations often arise during replication and packaging due to oversized constructs, DNA secondary structures, or processing errors. These incomplete genomes are often underreported by conventional analytical methods, diluting potency, inflating dose requirements, and driving up production costs.
Traditional analytical methods often fail to detect these incomplete genomes, masking the true ratio of functional to non-functional vectors. As a result, developers operate with incomplete information, leading to inefficiencies across process development, quality control, and scale-up.
Recent studies highlight the scale of the problem. McColl-Carboni et al. (Gene Therapy, 2024)found that many of the rAAV capsids presumed to be “full” actually carry truncated genomes that are unable to deliver therapeutic benefits. Similarly, Jiang and Dalby (Trends in Biotechnology, 2023) identified limitations incurrent analytics as a major barrier to scalable rAAV manufacturing.
There is a clear need for accurate, high-resolution quantification of genome integrity. Without it, gene therapy programs risk suboptimal outcomes at every stage of development.
The limitations of existing rAAV characterization techniques
Current tools for rAAV quantification serve various purposes, but they cannot directly assess transgene integrity. Whether based on optical properties, density gradients, or partial sequence detection, these methods lack the specificity needed to confirm full genomes.
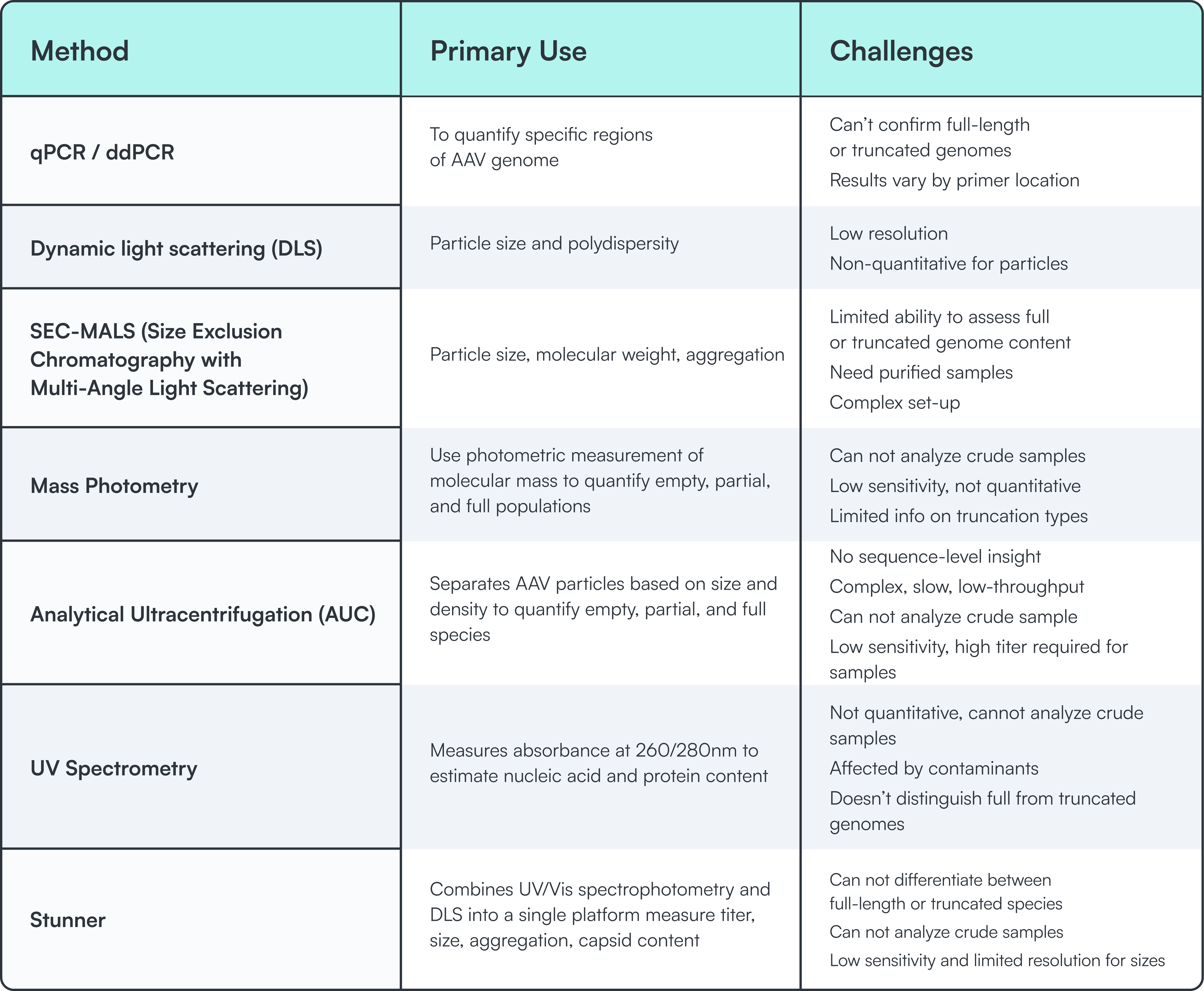
None of these approaches provide a direct, end-to-end measurement of transgene presence. Most require purified samples and involve complex workflows unsuitable for early-stage process development or high-throughput environments.
Download our white paper to see a more detailed comparison of various rAAV analytical approaches across various manufacturing steps. (here)
NanoMosaic:Molecular-level quantification of true full rAAV genomes
To address the critical gap in full-genome detection, NanoMosaic offers a nanoneedle-based affinity assay that directly quantifies full-length rAAV genomes with high specificity. It uses two capture probes targeting both ends of the transgene, ensuring signal is only generated when both termini are present, i.e. the complete genome. This allows true full genomes to be distinguished from truncated or partial forms with high specificity at a molecular level.
Key NanoMosaic Advantage
- Sequence-specific and structure-dependent: Only detects full-length genomes.
- Crude sample compatibility: Enables direct analysis of lysates and process intermediates without purification; allows in-process monitoring.
- Low sample volume: Requires only 2 μL per well; scalable across 96-, 384-, and 1536-well formats.
- High sensitivity and specificity: 20,000 nanoneedles per well for low-abundance detection.
- Scalable and Rapid: Streamlined, benchtop-friendly workflows ideal for process development, screening and QC.
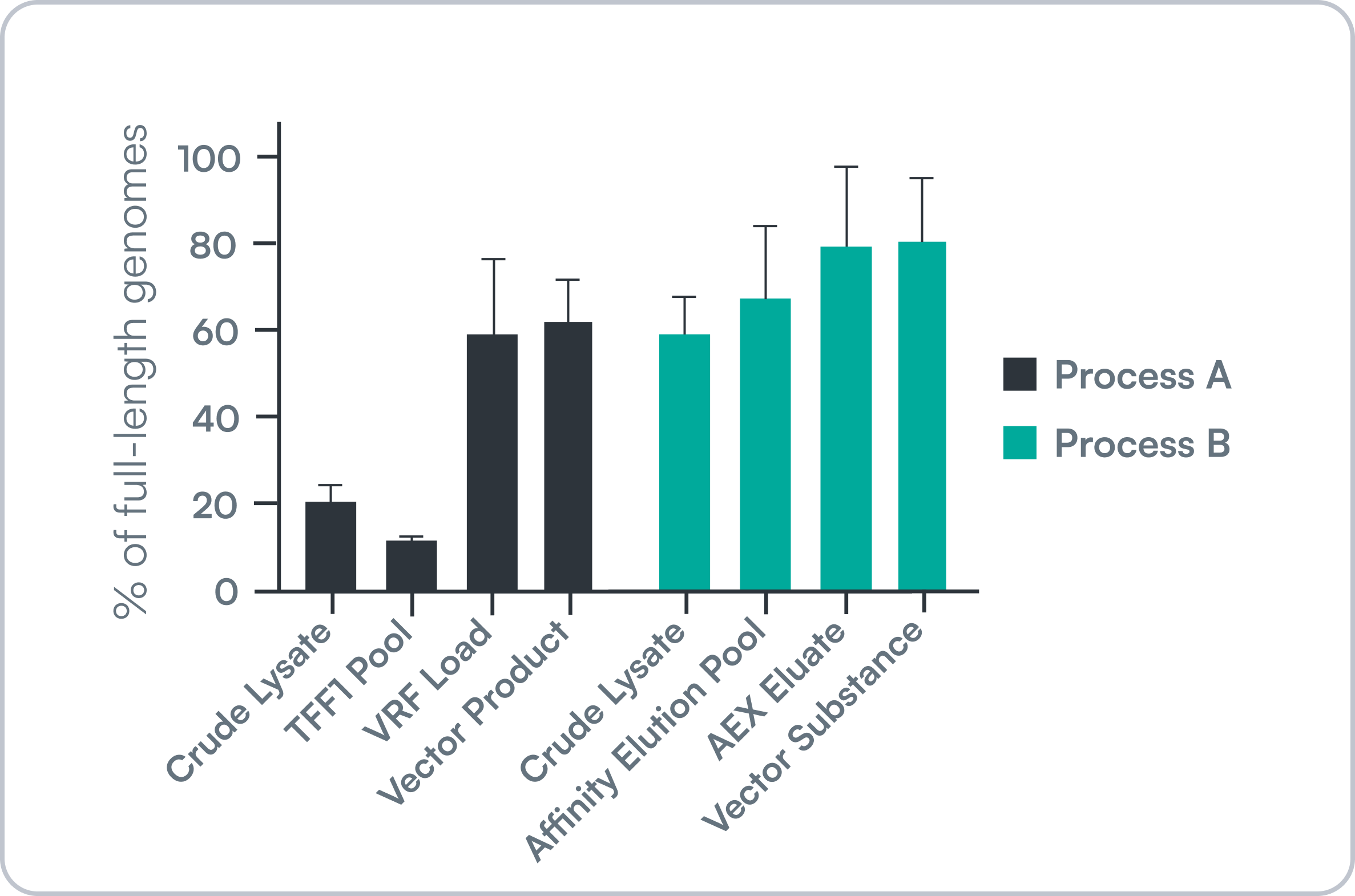
- Supports process optimization: Track full vs. truncated genomes from crude lysate to final substance to inform yield and quality improvements.
NanoMosaic distinguishes between truncated and full genomes at every stage of manufacturing, enabling more meaningful process optimization and QC decisions.
Still not sure which approach is best for your application?
Talk to us about integrating NanoMosaic into your analytics workflow.
Conclusion
As gene therapy programs mature, so must the analytics that support them. Rapid advancements in rAAV genome technology mean that the limitations of traditional methods threaten to compromise potency, scalability, and safety. NanoMosaic resolves this by providing high-resolution, sequence-dependent quantification that is fast, scalable, and robust across sample types.
Critically, NanoMosaic eliminates the need for sample purification, thereby enabling accurate genome quantification directly from crude lysates and early-stage intermediates. This unlocks earlier decision-making opportunities in processdevelopment, shortens feedback loops, and reduces the risk of scaling inefficiencies. By minimizing sample requirements and turnaround time, the platform supports a more agile, data-driven manufacturing strategy.
Accurateanalytics isn't just a QC tool—it’s a lever for reducing cost, improving yield,and accelerating therapeutic progress.
Unlock better process decisions, lower manufacturing risk, and increase vector quality.
Download our white paper for a full assessment of our nanoneedle approach, including:
- A direct side-by-side evaluation across multiple analytical platforms
- Genome integrity tracking from crude lysate to final drug substance
- Actionable insights for improving AAV vector quality and manufacturing efficiency
Ready to optimize your rAAV analytics? Let’stalk.
Advancing AAV analytics: filling a critical gap in full-length transgene detection

Recombinant adeno-associated viruses (rAAVs) are fundamental to gene therapy, but a persistent challenge limits their full potential: most viral particles lack the full, intact therapeutic payload. Truncations often arise during replication and packaging due to oversized constructs, DNA secondary structures, or processing errors. These incomplete genomes are often underreported by conventional analytical methods, diluting potency, inflating dose requirements, and driving up production costs.
Traditional analytical methods often fail to detect these incomplete genomes, masking the true ratio of functional to non-functional vectors. As a result, developers operate with incomplete information, leading to inefficiencies across process development, quality control, and scale-up.
Recent studies highlight the scale of the problem. McColl-Carboni et al. (Gene Therapy, 2024)found that many of the rAAV capsids presumed to be “full” actually carry truncated genomes that are unable to deliver therapeutic benefits. Similarly, Jiang and Dalby (Trends in Biotechnology, 2023) identified limitations incurrent analytics as a major barrier to scalable rAAV manufacturing.
There is a clear need for accurate, high-resolution quantification of genome integrity. Without it, gene therapy programs risk suboptimal outcomes at every stage of development.
The limitations of existing rAAV characterization techniques
Current tools for rAAV quantification serve various purposes, but they cannot directly assess transgene integrity. Whether based on optical properties, density gradients, or partial sequence detection, these methods lack the specificity needed to confirm full genomes.

None of these approaches provide a direct, end-to-end measurement of transgene presence. Most require purified samples and involve complex workflows unsuitable for early-stage process development or high-throughput environments.
Download our white paper to see a more detailed comparison of various rAAV analytical approaches across various manufacturing steps. (here)
NanoMosaic:Molecular-level quantification of true full rAAV genomes
To address the critical gap in full-genome detection, NanoMosaic offers a nanoneedle-based affinity assay that directly quantifies full-length rAAV genomes with high specificity. It uses two capture probes targeting both ends of the transgene, ensuring signal is only generated when both termini are present, i.e. the complete genome. This allows true full genomes to be distinguished from truncated or partial forms with high specificity at a molecular level.
Key NanoMosaic Advantage
- Sequence-specific and structure-dependent: Only detects full-length genomes.
- Crude sample compatibility: Enables direct analysis of lysates and process intermediates without purification; allows in-process monitoring.
- Low sample volume: Requires only 2 μL per well; scalable across 96-, 384-, and 1536-well formats.
- High sensitivity and specificity: 20,000 nanoneedles per well for low-abundance detection.
- Scalable and Rapid: Streamlined, benchtop-friendly workflows ideal for process development, screening and QC.
- Supports process optimization: Track full vs. truncated genomes from crude lysate to final substance to inform yield and quality improvements.
NanoMosaic distinguishes between truncated and full genomes at every stage of manufacturing, enabling more meaningful process optimization and QC decisions.

Still not sure which approach is best for your application?
Talk to us about integrating NanoMosaic into your analytics workflow.
Conclusion
As gene therapy programs mature, so must the analytics that support them. Rapid advancements in rAAV genome technology mean that the limitations of traditional methods threaten to compromise potency, scalability, and safety. NanoMosaic resolves this by providing high-resolution, sequence-dependent quantification that is fast, scalable, and robust across sample types.
Critically, NanoMosaic eliminates the need for sample purification, thereby enabling accurate genome quantification directly from crude lysates and early-stage intermediates. This unlocks earlier decision-making opportunities in processdevelopment, shortens feedback loops, and reduces the risk of scaling inefficiencies. By minimizing sample requirements and turnaround time, the platform supports a more agile, data-driven manufacturing strategy.
Accurateanalytics isn't just a QC tool—it’s a lever for reducing cost, improving yield,and accelerating therapeutic progress.
Unlock better process decisions, lower manufacturing risk, and increase vector quality.
Download our white paper for a full assessment of our nanoneedle approach, including:
- A direct side-by-side evaluation across multiple analytical platforms
- Genome integrity tracking from crude lysate to final drug substance
- Actionable insights for improving AAV vector quality and manufacturing efficiency
Ready to optimize your rAAV analytics? Let’stalk.
